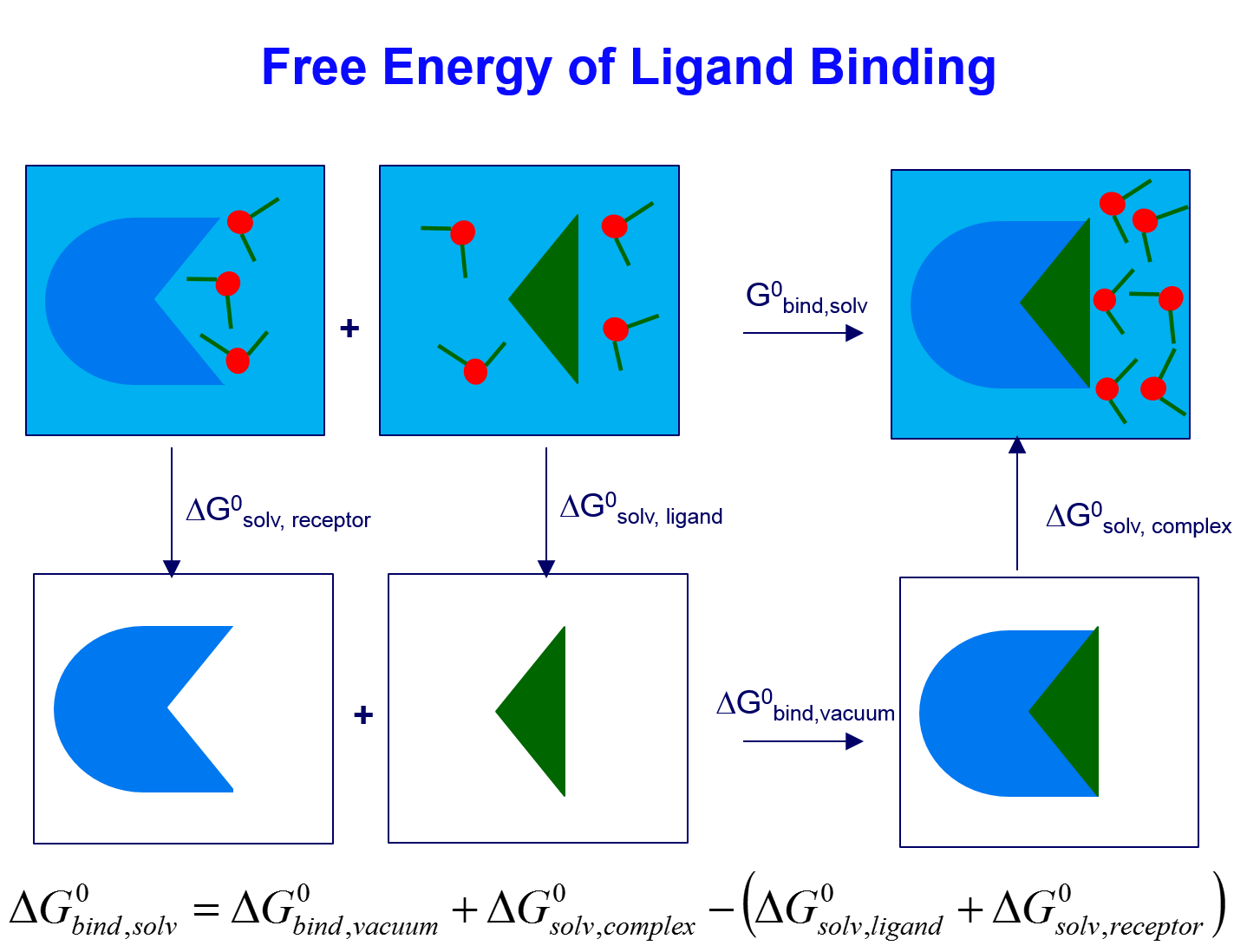
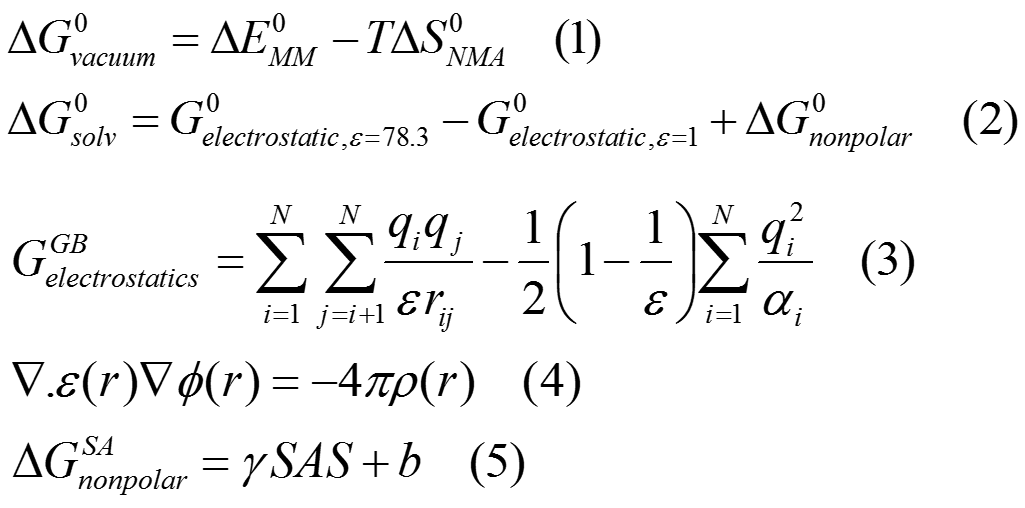In molecular docking, scoring functions are fast approximate mathematical methods used to predict the strength of the non-covalent interaction (also referred to as binding affinity) between the two molecules after the ligand (usually a small organic molecule) have been docked to the receptor (such as protein and DNA).
A scoring function in molecular docking falls into one of the three categories: molecular mechanical force field (MMFF)-based, empirical-based or knowledge-based. We are interesting to develop high quality MMFF-based scoring functions to accurately predict the binding affinity, not only considering the van der Waals and electrostatic interactions, but also the solvation contribution. The desolvation energies of the ligand and of the protein are taken into account by using implicit solvation methods of GBSA and PBSA.
How to Calculate Binding Free Energy Using MM-PB/GBSA
Principle of MM-PB/GBSA